Inheritance: Regulation of Gene Expression in Prokaryotes and Eukaryotes
Regulation of Gene Expression in Prokaryotes and Eukaryotes
Clever mechanisms turn genes off and on so that they only function when there is a need for their services.
Prokaryotes and the Operon Model
Prokaryotes are sensitive to their environment, and their genetic activity is controlled by specific proteins that interact directly with their DNA to quickly adjust to environmental changes. Genetic expression is the process where genotypes coded in the genes are exhibited by the phenotypes of the individuals. The DNA is copied by the RNA and then synthesized into protein. The process of transcription, which is the synthesis of RNA from a DNA template, is where the regulation of the gene expression is most likely to occur. The default setting for prokaryotes appears to allow for the continual synthesis of protein to occur, whereas in eukaryotes the system is normally off until activated.
An operon is a self-regulating series of genes that work in concert. An operon includes a special segment of genes that are regulators of the protein synthesis, but do not code for protein, called the promoter and operator. These segments overlap, and their interaction determines whether the process will start and when it will stop. RNA polymerase must create RNA by moving along the chromosome and “reading” the genes in the process of transcription.
RNA polymerase first attaches to the promoter segment, which signals the beginning of a particular DNA sequence. If not blocked, it passes over the operator and reaches the protein-producing genes where it creates the mRNA that instructs the ribosomes to create the desired protein. This process continues until the system is blocked by repressor proteins. Repressors bind with the operator and prevent RNA polymerase from proceeding to create mRNA by prohibiting access to the remainder of the protein-producing genes. As long as the repressor is binding with the operator, no proteins are made. However, when an inducer is present, it binds with the repressor, causing the repressor to change shape and release from the operator. When this happens, the RNA polymerase can proceed with transcription, and protein synthesis begins and continues until another repressor binds with the operator. Refer to the illustration Transcription regulation.
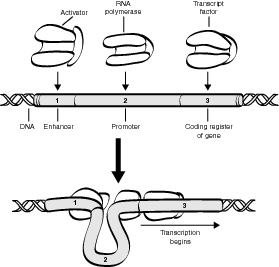
The lac operon model is probably the most studied and well known. In bacteria, such as E. coli, three genes are part of an operon that code for three separate enzymes needed for the breakdown of lactose, a simple sugar. A regulatory gene, located before the operon, continually makes repressor proteins that bind with the operator and prohibit the function of RNA polymerase. The system therefore remains off until a flood of lactose molecules binds with all available repressors and prevents their attachment to the operator. When the operator is free, the production of the enzyme to break down lactose continues until enough of the lactose molecules are broken down to then release repressors to recombine with the operator to stop production of the enzymes.
Two additional types of operons exist that operate in the same way except for the function of the operator. The trp operon differs because the repressor is active only when bonded to a specific molecule. For the remainder of the time, it remains unbonded and inactive in the absence of that molecule. Finally, in a positive twist, activators are used by a third type of operon to bond directly with the DNA, which allows the RNA polymerase to work more efficiently. Absent the activators, RNA polymerase proceeds at a slow rate.
Eukaryotes: Multiple Models of Gene Regulation
Unlike prokaryotes, multiple gene-regulating mechanisms operate in the nucleus before and after RNA transcription, and in the cytoplasm both before and after translation.
Histones are small proteins packed inside the molecular structure of the DNA double helix. Tight histone packing prevents RNA polymerase from contacting and transcribing the DNA. This type of overall control of protein synthesis is regulated by genes that control the packing density of histones. X-chromosome inactivation occurs when dense packing of the X chromosome in females totally prevents its function even in interphase. This type of inactivation is inherited and begins during embryonic development, where one of the X chromosomes is randomly packed, making it inactive for life.
Activator-enhancer complex is unique in eukaryotes because they normally have to be activated to begin protein synthesis, which requires the use of transcription factors and RNA polymerase. In general, the process of eukaryotic protein synthesis involves four steps:
- Activators, a special type of transcription factor, bind to enhancers, which are discrete DNA units located at varying points along the chromosome.
- The activator-enhancer complex bends the DNA molecule so that additional transcription factors have better access to bonding sites on the operator.
- The bonding of additional transcription factors to the operator allows greater access by the RNA polymerase, which then begins the process of transcription.
- Silencers are a type of repressor protein that blocks transcription at this point by bonding with particular DNA nucleotide sequences.
The processing and packaging of RNA both in the nucleus and cytoplasm provides two more opportunities for gene regulation to occur after transcription but before translation.
Adding extra nucleotides as a protective cap and tail to the RNA identifies the RNA as an mRNA by the ribosomes, and prevents degradation by cell enzymes as it moves from the nucleus into the cytoplasm.
RNA splicing occurs when “gaps” of nonprotein-code-carrying nucleotides called interons are removed from the code-carrying nucleotides, called exons, which are then connected to shorten the RNA molecule for conversion into tRNA and rRNA. The number of interons regulates the speed at which the RNA can be processed.
After the extra nucleotides have been added as a cap and tail and the RNA has been spliced, it moves to the cytoplasm where additional mechanisms of gene regulation exist.
The longevity of the individual mRNA molecule determines how many times it can be used and reused to create proteins. In eukaryotes, the mRNA tends to be stable, which means it can be used multiple times; which is efficient, but it prevents eukaryotes from making rapid response changes to environmental disruptions. The mRNA of prokaryotes is unstable, allowing for the creation of new mRNA, which has more opportunities to adjust for changing environmental conditions.
Inhibitory proteins prevent the translation of mRNA. They are made inactive when bonded with the substance for which they are trying to block production.
Post-translation control involves the selective cutting and breakdown of proteins that prevent the formation of the final product. In both cases, the hormone or enzyme required to finish or activate the final product may be rendered inactive.
Although much has been learned about inheritance since Mendel's time, the fundamentals remain the same. In sexual reproduction the offspring inherit one half of their genes from the father and one half from the mother. The chance of inheriting a particular gene can be estimated by pedigrees and Punnet squares. Every trait, feature, or characteristic is controlled by genes or a combination of genes. Numerous gene-regulating mechanisms activate and inactivate organism functions.
Excerpted from The Complete Idiot's Guide to Biology © 2004 by Glen E. Moulton, Ed.D.. All rights reserved including the right of reproduction in whole or in part in any form. Used by arrangement with Alpha Books, a member of Penguin Group (USA) Inc.
To order this book direct from the publisher, visit the Penguin USA website or call 1-800-253-6476. You can also purchase this book at Amazon.com and Barnes & Noble.







